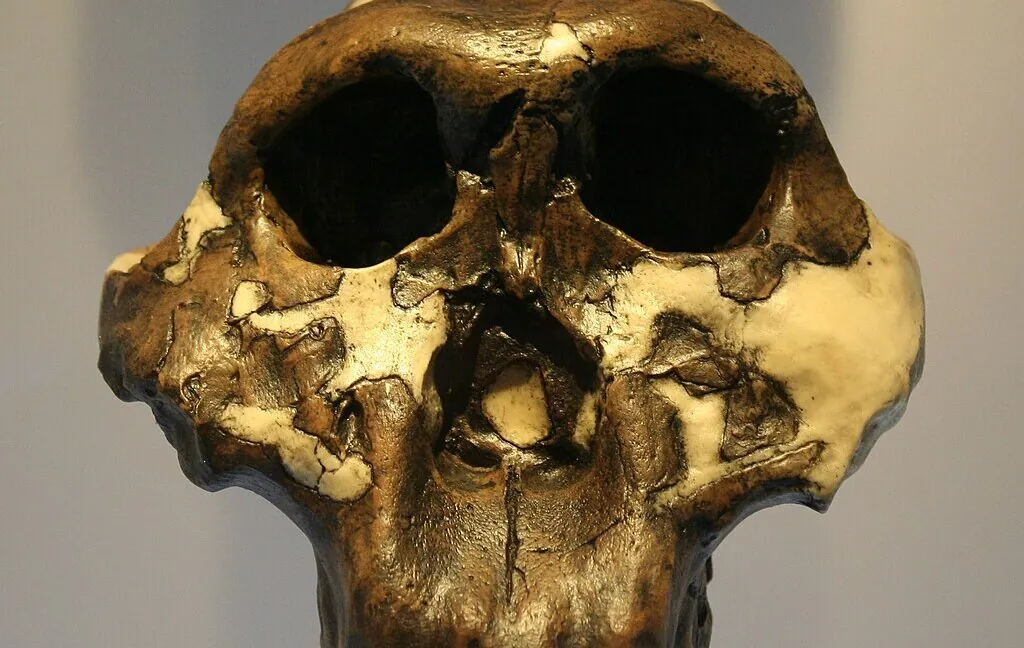
They’ve extracted fragments of enamel proteins from the teeth of fossils of the species Paranthropus robustus and used them to test whether the remains truly belonged to one species, despite dramatic differences in size.
A complicated species Remains that have been classified as Paranthropus show up in the fossil record nearly 3 million years ago and persist for roughly a million years.
So, for Paranthropus remains that date from roughly 2 million years ago, the team turned to proteins found in teeth.
Using this, the researchers could identify the likely composition of individual fragments of proteins and match those to known tooth enamel proteins.
Among the 425 different amino acids were 16 that had species-specific variations among hominins.
Ancient DNA research has completely changed our understanding of the history of our own species. It has indicated the existence of Denisovans and clarified our relationship with Neanderthals. However, DNA deteriorates over time, even in the best conditions, so there is a limit to how far back we can hope to answer questions about our ancestors. Furthermore, Africa, where conditions are much less conducive to DNA’s survival, is home to the majority of the species that we have learned about.
A sizable international team has now discovered an alternative method for extracting some genetic information from much older remains. Despite the stark size differences, they have used pieces of enamel proteins that were taken from the teeth of fossils of the species Paranthropus robustus to determine whether the remains actually belonged to a single species. Because one of the proteins is specific to men, they also discovered that an individual’s size was not always correlated with its sex.
The species is complex.
Approximately 3 million years ago, remains identified as Paranthropus appeared in the fossil record and remained for about a million years. It thus shared similarities with early Homo genus members as well as australopithecines. This genus has been assigned four distinct species, but the situation is complex. Because of its many similarities to certain Australopithecus species, interbreeding may be possible. Within remains that have been identified as Paranthropus, there is also a great deal of variation, particularly in the size of individuals. It has proven challenging to test the theory that this could be the result of “sexual dimorphism,” or the differences between males and females in this species.
Regretfully, no remains have been discovered outside of Africa, and no DNA from that continent that is older than 20,000 years—long after Paranthropus went extinct—has been recovered. In order to find remains of Paranthropus that are approximately 2 million years old, the team looked to tooth proteins. Over time, these will unavoidably deteriorate, breaking down into smaller pieces and undergoing chemical changes to certain amino acids. However, tooth enamel’s resilience should guard against their complete devastation.
Additionally, the mass spectrometry technique is sensitive enough to detect individual atoms’ isotopes combined with larger molecules. By doing so, the scientists were able to determine the probable makeup of distinct protein fragments and compare them to proteins found in tooth enamel.
The researchers tested their methods on animal remains discovered with the hominins before using samples of Paranthropus teeth, which were all discovered at the same location within the Cradle of Humankind World Heritage Site in South Africa. Even though the proteins found differed from sample to sample, they discovered six distinct proteins that were present in all four of the teeth under study—albeit only in fragments. 425 amino acids from the six proteins were covered by those fragments taken together.
In order to verify the authenticity of the results produced in a Copenhagen lab, they replicated the procedure at a Cape Town lab. Some of the amino acids displayed indications of chemical damage, suggesting that they were, in fact, fragments of proteins that were two million years old. This helped to support the successful replication.
reading the amino acids.
What can we learn from tiny fragments of ancient proteins? More than you might think. The reason for this is that a gene on the Y chromosome produces the enamel protein AMELY (the X chromosome also contains the AMELX gene). Furthermore, it is evident that some of the protein fragments found in a subset of the four remaining pieces of AMELY were part of the tooth, suggesting that the person who had it was male.
For testing the hypothesis that body size is a measure of sex differences in Paranthropus, that proves to be crucial. One of the samples was a tiny tooth that had been assumed to belong to a woman, but it is clearly identified as male due to the presence of AMELY. In this species, “it enables us to exclude sexual dimorphism as one of the multiple variables affecting the range of anatomical variation,” according to the researchers, making it a significant finding.
A sample may be female if AMELY is absent, but this is not a guarantee. This is partly due to the fact that some rare males (including at least one Neanderthal) carry deletions that completely eliminate the gene, and partly because it is impossible to rule out some issue with identifying the protein in samples this old.
The fact that some of the 425 amino acid locations vary among hominin species and even among individual Paranthropus species is another important consideration. Therefore, they may be able to help clarify some of the ambiguity surrounding the number of Paranthropus species and their relationships with other hominins, as well as act as a diagnostic tool for relationships both within and between species. Even though four samples isn’t enough to say much, the researchers discovered some encouraging data.
For instance, they investigated whether the type of variation in amino acids observed in these samples could be observed if they were all members of the same species. To do this, four human genomes were selected at random, and their levels of variation were compared. Although they came to the conclusion that it was “plausible” that four randomly selected individuals would exhibit this degree of variation, the test was not conclusive because the population of modern humans is probably larger than that of Paranthropus.
Of the 425 amino acids, 16 were species-specific for hominins. According to a tree constructed from these variations, Paranthropus robustus is, somewhat surprisingly, the species most closely related to our own genus, Homo. But again, they come to the conclusion that there is just not enough information to be certain of this conclusion.
However, that ought to be a “not enough data yet” statement. Regular Ars reader Enrico Cappellini, who is also the paper’s senior author and a faculty member at the Globe Institute at the University of Copenhagen, told us about it. Additionally, his primary research focus, according to his faculty profile, is improving the methods employed here, so perhaps in the future we’ll be able to increase the amount of information on extinct hominin species. As the paper points out, the technique’s drawback is that it only destroys a tiny portion of the sample, which are unique fragments of humanity’s collective history.
Science. 2025. Regarding DOIs, the DOI is 10.1126/science . adt9539.